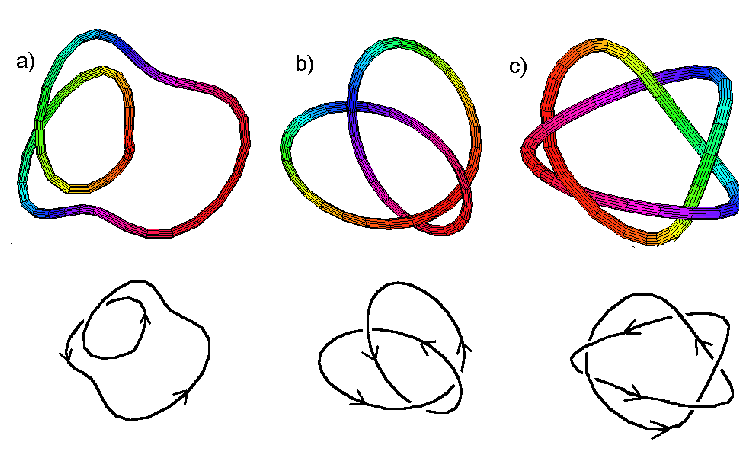
Introduction: DNA is the genetic material of all cells, containing coded information about cellular molecules and processes. DNA consists of two polynucleotide strands twisted around each other in a double helix. The first step in cellular division is to replicate DNA so that copies can be distributed to daughter cells. Additionally, DNA is involved in transcribing proteins that direct cell growth and activities. However, DNA is tightly packed into genes and chromosomes. In order for replication or transcription to take place, DNA must first unpack itself so that it can interact with enzymes.
DNA packing can be visualized as two very long strands that have been intertwined millions of times, tied into knots, and subjected to successive coiling. However, replication and transcription are much easier to accomplish if the DNA is neatly arranged rather than tangled up in knots. Enzymes are essential to unpacking DNA. Enzymes act to slice through individual knots and reconnect strands in a more orderly way.
Importance: We can gain insight into the unknotting of DNA by using principles of topology. Topologists study the invariant properties of geometric objects, such as knots. Tightly packed DNA in the genes must quickly unknot itself in order for replication or transcription to occur. This is a topological problem.
Question: How can knot theory help us understand DNA packing? How can we estimate the rates at which enzymes unknot DNA?
Methods: DNA can be visualized as a complicated knot that must be unknotted by enzymes in order for replication or transcription to occur. It is perhaps not surprising then that connections between mathematical knot theory and biology have been discovered. By thinking of DNA as a knot, we can use knot theory to estimate how hard DNA is to unknot. This can help us estimate properties of the enzymes that unknot DNA.
A mathematical knot is a closed curve. This can be visualized as a closed loop of string. If the string had a knot in it, it would be impossible to unknot without slicing throught the knot. Look at the pictures below.
The first knot is merely a loop of string that has been twisted, an "unknot". It could easily be unknotted by pulling on the string to form a single loop. The 2nd knot, however, is clearly a knot. The only way to get rid of the knot would be to cut through it and retie the string. The 3rd knot is even more complicated.
To understand this more rigorously, we will explore some topological definitions. A knot is a closed curve in three-dimensional space. Two knots are considered the same if one can be moved smoothly through space, without any cutting, so that it is identical to the second. Mathematical knots are represented by two-dimensional diagrams that can be thought of as the shadow cast by a three-dimensional knot.
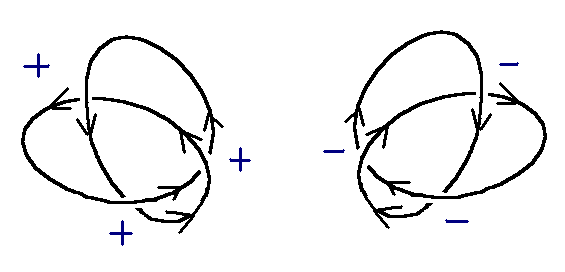
At points in the diagram, the curve will cross over or under itself. The crossing points are called doublepoints. Each double point is assigned a + or - sign, depending on the orientation of the crossing point. If the strand passing over a doublepoint can be turned clockwise less than 180 to match the direction of the strand underneath, then the sign is positive (+); if the strand on top must be rotated counter-clockwise, it is negative (-). The writhe of a knot is the sum of all signs of its doublepoints. The knots below are +3 and -3, respectively.
The only way to untie a mathematical knot is to cut through the knot so that the strand that was lying on top is now underneath. This is equivalent to changing the sign of a doublepoint. The number of times on must allow one strand of a knot to pass through another (in order to unknot it), is called the unknotting number. If a knot can be moved smoothly through three-dimensional space to remove the doublepoint, then that doublepoint does not count toward the unknotting number. Consequently, in the figure below, knot a) has an unknotting number of zero, despite having one crossing point. Knot b) has an unknotting number of 1, despite having 3 crossing points.
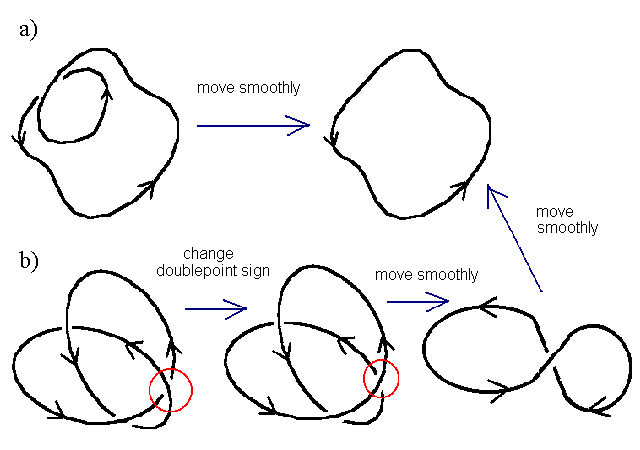
A knot is in its ideal form if it has been moved smoothly through space so that all excess doublepoints are removed. Knots are generally described by two numbers, CU, the ideal crossing number (C) and the unknotting number (U). In the figure above, the knots would be 00 and 31, respectively. Mathematical knots may have values for writhe and crossing number that are much higher than the ideal number if they have been moved smoothly through space to a more complicated form.
Interpretation: We can extend the principles of topology to DNA. The value of this method is that it gives researchers a quantitative measure for DNA packing, rather than a subjective description. In DNA packing and unpacking, an enzymatic reaction converts DNA strands to knots, linked strands, or a more orderly form. Knot theory can help scientists discover the mechanisms by which these enzymes work.
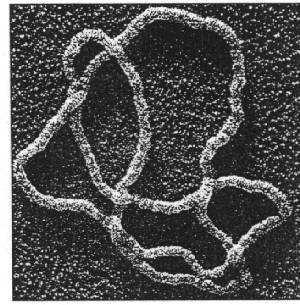
Picture taken from Sumners, D. 1995. Lifting the curtain: Using topology to probe the hidden action of enzymes. Notices of the AMS 42:528-537.
Scientists use electron microscopes to take pictures of DNA. Underlying and overlying segments are distinguished by using a protein coating. The flattened DNA is then visualized as a knot. The unknotting number and ideal crossing number can then be estimated. For several DNA fragments with the same knot number, CU, there may be a variety of different forms as the DNA is twisted and distorted. However, the average writhe and crossing number can be estimated for a particular ideal knot number.
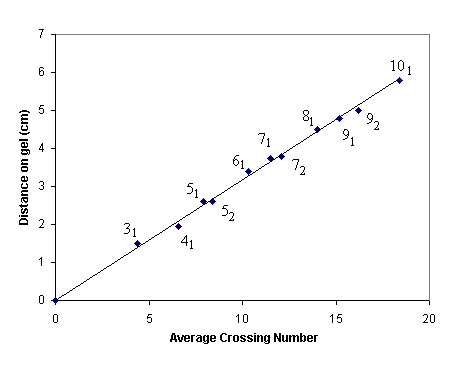
A more convenient procedure for determining the crossing number of DNA knots involves using gel electophoresis. The distance DNA fragments move on an electrophoretic gel is highly correlated with the average crossing number (see graph). The ideal knot number is given on the graph (reproduced from Stasiak et al 1996).
Using topological methods helps scientists gain insight into properties of DNA replication, transcription, and recombination. Enzymes called topoisomerases are important in DNA unpacking and packing. In addition to its double-helix structure, DNA has a number of twists in it as it coils around itself called supercoiling. Brown and Cozzarelli (1979) used topological methods to determine how the topoisomerase, gyrase, works in E. coli to supercoil DNA. If we represent this as a 2-D drawing, we can count the number of doublepoints and determine their signs.
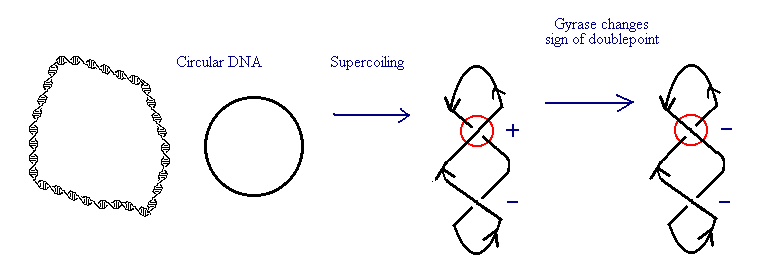
Brown and Cozzarelli found that the writhe decreased in a stepwise fashion as gyrase acted. A stepwise decrease in writhe would occur as the sign of each doublepoint changes from positive to negative. They concluded that topoisomerases act by slicing through each double point and reconnecting the strands on the reverse side.
Topological methods also distinguished two classes of topoisomerases. Brown and Cozzarelli found that the stepwise decrease in crossing number consisted of two steps. This indicated that gyrase was a type 2 topoisomerase that acted by nicking both strands of the double helix, and reconnecting them underneath. In contrast, type 1 topoisemerases unpack DNA by nicking a single strand, rotating the free ends to relieve the supercoiling, and reconnecting the ends. In this case, the stepwise decrease in crossing number would consist of a single step.
Topoiserases act locally, breaking apart a single doublepoint before they are free to move to another one. Computing the unknotting number for twisted DNA gives cell biologists an estimate of the number of times a topoisomerase must act in order to unpack the DNA. We can measures the reaction rates (number of strand crossings per minute per mole of enzyme) for particular enzymes (See Enzymes and the Rates of Chemical Reactions.). In this case, the crossing points of the DNA or DNA of a particular knot type would be the substrate. An enzyme solution is added to the DNA solution. Product is formed as the enzymes change the signs of doublepoints or create different knot types. Measuring changes in topology to estimate reaction rates gives us a quantitative measure for comparing different enzymes or a single enzyme under different conditions.
Conclusions: Principles of topology give cell biologists a quantitative, powerful, and invariant way to measure properties of DNA. Principles of knot theory have helped elucidate the mechanisms by which enzymes unpack DNA. Additionally, topological methods have been influential in determining the left handed winding of DNA around histones. Measuring changes in crossing number have also been instrumental in understanding the termination of DNA replication and the role of enzymes in recombination.
Additional Questions:
1. Calculate the writhe, number of doublepoints, and the uncrossing number for knot c) in the first figure.
Sources:
Brown, P. O. and N. R. Cozzarelli. 1979. A sign inversion mechanism for enzymatic supercoiling of DNA. Science 206:1081-1083.
Menasco, W. and L. Rudolph. 1995. How hard is it to untie a knot? American Scientist 83:38-49.
Stasiak, A., V. Katritch, J. Bednar, D. Michoud, and J. Dubochet. 1996. Electrophoretic mobility of DNA knots. Nature 384:122.
Wasserman, S. A. and N. R. Cozzarelli. 1986. Biochemical topology: Applications
to DNA recombination and replication. Science 232:951-960.
Copyright 1999 M. Beals, L. Gross, S. Harrell